M/C Lineage Admixture Assay – Single Colony
£55.00
Description
This test analyses the level of hybridisation present in the genetics of a single queen, by analysing the DNA of her drones.
For more information on why you might choose this test, and how it compares to our other services, see our Services page.
The price of this test includes the cost of shipping of the sample kit, and returning the samples to Beebytes.
An example report, showing how the results from this assay are presented, can be found under the ‘Downloads’ Tab for this product.
The sampling instructions are included in the kit, but can also be found under the ‘Downloads’ Tab for this product.
Science
The iPLEX MassARRAY developed by Henriques et al. (2018) and our very own Dr Mark Barnett. This platform is typically used on DNA extracted from drones enabling the queen’s genotypes at these SNPs to be inferred. The platform is specifically designed to test for C lineage admixture levels in M lineage honey bees. We have compared admixture estimates for drones we have whole genome sequence (WGS) data for to estimates based on this array, and the results are very consistent.
The test analyses a small number of variants in the bees’ DNA (known as single nucleotide polymorphisms or ‘SNPs’). These particular variants have been shown to differentiate the 2 main honey bee lineages present in the UK: M – mellifera, or C – carnica/ligustica. This test allows to see how much hybridisation (admixture) is present in the bees, and is useful for projects focusing on breeding ‘native’ bees, selecting for M or C lineage. Traditionally native bees were identified based on physical characteristics such as wing venation and hair colour, however these will be under the genetic control of a very small number of the 10,000+ genes in the genome. Consequently, breeding on these characteristics alone will simply select for the handful of genes involved in their development. The SNPs on this however are distributed throughout the genome, providing a much more balanced and informed approach.
The Mitochondrial DNA assay uses PCR amplification and Sanger sequencing of tRNA-COII mitochondrial barcode using E2 and H2 primers designed by Garnery et al. (1992). This method will indicate maternal ancestry from the 16 kb mitochondrial genome only.
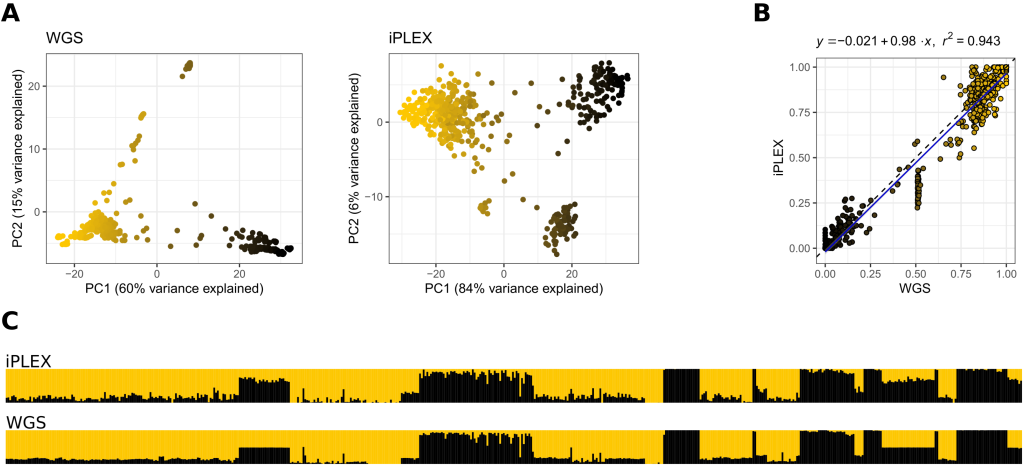
A Principal Components Analysis (PCA) reveals the same clusters in WGS and iPLEX data. B C-lineage ancestry estimates are highly correlated (r2 = 0.94) between WGS and iPLEX data. C Admixture (ALStructure) plots for WGS and iPLEX data assuming 2 genetic backgrounds.
Only logged in customers who have purchased this product may leave a review.




Reviews
There are no reviews yet.